1. Impacts of public-health interventions on infectious disease transmission
|
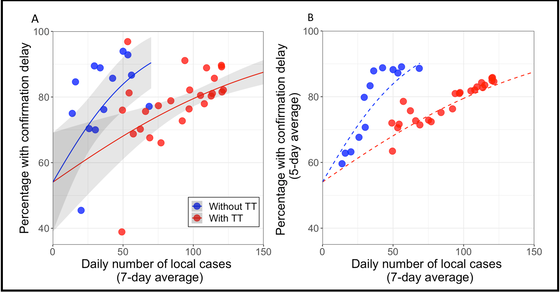
Figure 1. Percentage of epi-linked cases with confirmation delay as a function of the change in daily number of cases (average by week), before (blue) and after (red) the introduction of targeted group testing. (Lancet Regional Health 2022(i)).
|
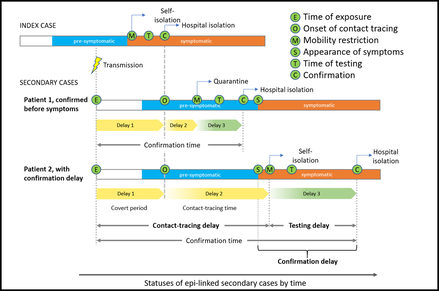
Figure 2. Chematic of the delays in contact tracing, testing and confirmation for epi‑linked cases. (Lancet Regional Health 2022(ii)).
|
|
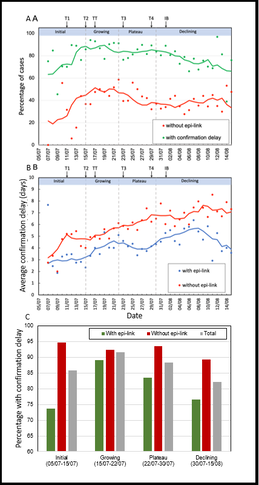
Our group has previously worked with an Oxford Professor, Colin BLAKEMORE, to develop a methodology to study the efficiency of contact tracing. We identified percentage of confirmation delay among contact-traced cases is an important indicator of contact-tracing efficiency in Hong Kong (refer to figures 1, 3). Then we incorporated such delays in our modelling in order to quantify the impacts of each major intervention (refer to figure 2).
Our group has been currently working on a risk assessment of current COVID-19 by modelling the impacts of border controls and other public health measures using a meta-population model. During the SARS-CoV-2 early spreading period, we have developed an “easy-to-use” mathematical framework to determine the probability of community spread. Using the top 10 visiting cities from Wuhan in China as an example, we first demonstrated that the arrival time and the dynamics of the outbreaks at these cities can be successfully predicted under the reproduction number R0 = 2.92 and incubation period τ = 5.2 days. The work has been published in Epidemics 2020. |
2. Impacts of climate change on weather variation on infectious disease transmission
Our group has found that a correlation exists between the weather and humidity that patients are exposed to during different stages of COVID-19 infection and the probability of death. The results show that weather conditions were associated with fatality rates among COVID-19 cases differently, i.e. during different stages of infection (Environmental Research 2022).
|
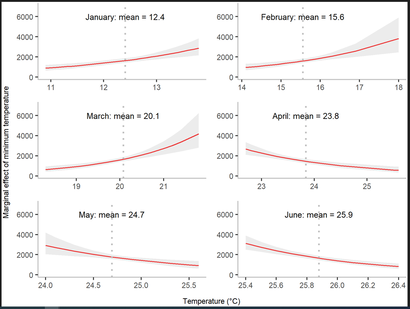
Figure 4. Marginal effect of minimum temperature from January to June on annual dengue cases. (PLOS Global Public Health 2022)
|
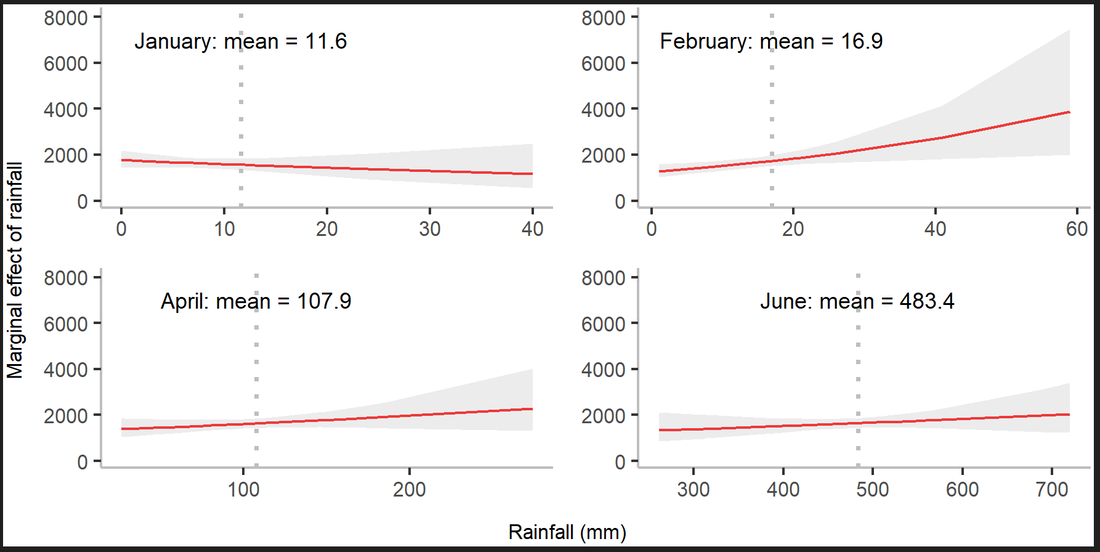
Figure 5. Marginal effect of total rainfall on annual dengue incidence in January, February, April and June. (PLOS Global Public Health 2022)
|
Our group also focuses on modelling the impact of extreme weather conditions on Dengue virus transmission. Nonlinear effects of both rainfall and temperatures on mosquito population size exist.
Mathematical modelling of mosquito population size would improve the accuracy of forecasting Dengue incidence. Understanding how climate variation drives Dengue outbreaks through mosquito population dynamics in East Pacific Asian areas (such as Hong Kong, Taiwan and Bangladesh) would help to understand whether Dengue infection is currently moving from tropical toward temperate areas and is of great importance to health protection not just in East Asia but also subtropical areas in other parts of the world. The work on Dengue forecast and control currently collaborates with National Health Research Institutes and National Taiwan University in Taiwan. We are also developing new models to identify the lagged effects of weather on mosquito activities in Hong Kong.
3. The interplay between the transmission of the virus and its evolutio
|
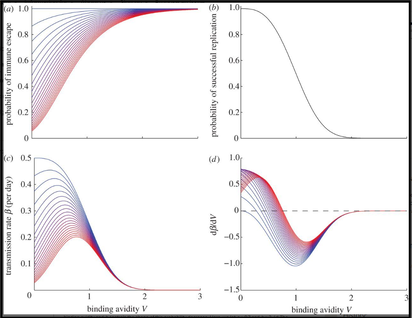
Figure 6. Effect of receptor binding avidity and previous exposure history on the transmission rate and its components. (The Royal Society 2013)
|
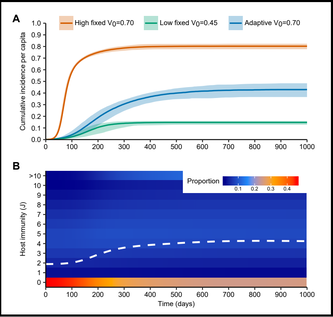
Figure 7. Dynamics of simulated epidemics with and without within-host binding avidity adaptation. (BioRxiv 2020)
|
One of our interests is to use mathematical modeling and evolutionary analysis to study the influenza viruses and host immunity dynamics in the population, including
i) how influenza viruses escape immunity
ii) how such viruses boost individual immunity in a population
iii) how binding avidity adaptation limits influenza transmission and diversity.
The study of the impact of influenza herd immunity and binding avidity collaborates with Prof. Steven Riley at Imperial College London, UK. We are also interested in studying the effects of social interactions on disease transmission. This is joint work with Prof. Kin On KWOK at CUHK.
i) how influenza viruses escape immunity
ii) how such viruses boost individual immunity in a population
iii) how binding avidity adaptation limits influenza transmission and diversity.
The study of the impact of influenza herd immunity and binding avidity collaborates with Prof. Steven Riley at Imperial College London, UK. We are also interested in studying the effects of social interactions on disease transmission. This is joint work with Prof. Kin On KWOK at CUHK.
Other research collaboration
We are also interested in applying mathematical modeling and computational approaches to study biological systems and to help experimentalists to simulate the impacts under different conditions to better understand biological systems. For example, we are currently using the Deep Learning technique to identify virulent pathways in Pseudomonas aeruginosa, which is an important cause of gram-negative infection, especially among immune-compromised patients in hospitals. We are also interested in building software tools to study therapeutic responses with next-generation sequencing techniques for precision medicine.
We are also interested in applying mathematical modeling and computational approaches to study biological systems and to help experimentalists to simulate the impacts under different conditions to better understand biological systems. For example, we are currently using the Deep Learning technique to identify virulent pathways in Pseudomonas aeruginosa, which is an important cause of gram-negative infection, especially among immune-compromised patients in hospitals. We are also interested in building software tools to study therapeutic responses with next-generation sequencing techniques for precision medicine.